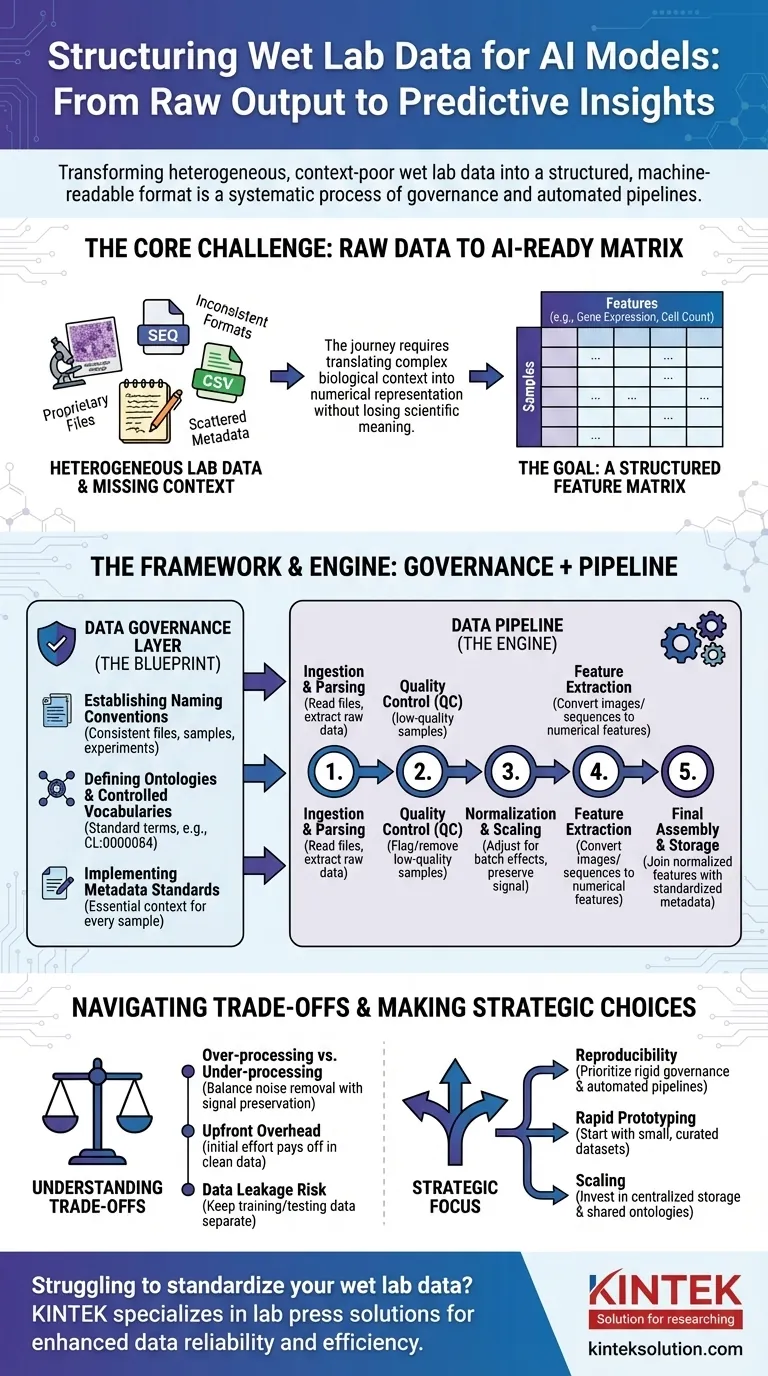
To prepare wet lab data for AI, you must transform it from its raw, often inconsistent state into a structured, machine-readable format. This is not a single step but a systematic process involving data governance to create clear rules, followed by data pipelines that automate the cleaning, normalization, and structuring of raw experimental outputs into a consistent format suitable for model training.
The core challenge is not simply reformatting files. It is about systematically translating complex biological context—such as experimental conditions, sample history, and measurement techniques—into a structured, numerical representation that an AI model can learn from without losing critical scientific meaning.

The Core Problem: From Raw Output to AI-Ready Data
The journey from a lab bench to a predictive model is fraught with data challenges. The raw output from scientific instruments is rarely, if ever, ready for direct use in an AI algorithm.
The Heterogeneity of Lab Data
Wet lab data comes in a vast array of formats. This includes everything from proprietary files from sequencers and microscopes to simple CSVs from plate readers, each with its own structure and quirks.
An AI model, however, requires a unified format.
The Curse of Missing Context
Critical information, or metadata, is often scattered. It might be in a scientist's notebook, a separate spreadsheet, or simply in their head. Without this context (e.g., which drug was applied, the temperature, the cell line used), the numerical data is meaningless.
The Goal: A Feature Matrix
Ultimately, most AI models need data in a feature matrix. This is a simple table where rows represent individual samples (e.g., a patient, a cell culture well) and columns represent features (e.g., gene expression levels, cell morphology measurements, protein concentrations).
A Framework for Standardization: The Data Governance Layer
Before you can build automated pipelines, you must establish rules. This is data governance—the blueprint that ensures consistency across all experiments and teams. It's the most critical and often overlooked step.
Establishing Naming Conventions
A simple but powerful rule is to enforce a consistent naming scheme for files, samples, and experiments. This allows data to be programmatically linked and tracked from its origin to the final analysis.
Defining Ontologies and Controlled Vocabularies
An ontology provides a standard set of terms for describing biological entities. For example, instead of allowing "T-cell," "T lymphocyte," and "Tcell," a controlled vocabulary enforces a single term, like CL:0000084 from the Cell Ontology.
This prevents ambiguity and ensures that data from different experiments is truly comparable.
Implementing Metadata Standards
You must define the minimum metadata that must be captured for every single sample. This often includes sample source, experimental conditions, instrument settings, and date. This rule ensures no data point becomes an orphan, detached from its context.
The Engine of Transformation: Building the Data Pipeline
With governance rules in place, you can build a data pipeline. This is a series of automated software steps that transforms raw data into the final AI-ready feature matrix.
Step 1: Data Ingestion and Parsing
The pipeline's first job is to find and read the raw data files. This step involves writing specific parsers for each instrument's output format to extract the primary measurements and any associated metadata.
Step 2: Quality Control (QC)
Not all data is good data. The pipeline should automatically flag or remove low-quality samples based on predefined metrics, such as low cell counts in an imaging experiment or poor read quality from a sequencer.
Step 3: Normalization and Scaling
Measurements from different batches or plates often have technical variations. Normalization is a crucial step that adjusts the data to make measurements comparable across experiments, removing technical noise while preserving biological signal.
Step 4: Feature Extraction
Raw data is often not in a feature format. An image, for example, must be processed to extract numerical features like cell size, shape, and intensity. A DNA sequence might be converted into a k-mer frequency vector. This step turns complex data into numbers the AI can use.
Step 5: Final Assembly and Storage
Finally, the pipeline joins the normalized features with the standardized metadata. This creates the final, clean feature matrix, which is then saved in a stable, queryable format (like Parquet or a database) for model training.
Understanding the Trade-offs
Structuring data is not a neutral process. Every choice you make can influence the final model's performance and interpretation.
Over-processing vs. Under-processing
Aggressive normalization or filtering can sometimes remove subtle but important biological signals. Conversely, failing to remove technical noise will guarantee your model learns from experimental artifacts instead of biology. This is a constant balance.
Standardization Creates Upfront Overhead
Implementing data governance requires significant initial effort and buy-in from the entire team. It can feel like it slows down research at first, but it pays massive dividends by preventing months of cleanup work later.
The Danger of Data Leakage
A critical pipeline function is to keep training and testing data separate. If information from the test set (e.g., its overall distribution) is used to normalize the training set, your model's performance will be artificially inflated and it will fail in the real world.
Making the Right Choice for Your Goal
Your approach to data structuring should be guided by your ultimate objective.
- If your primary focus is reproducibility: Prioritize rigid data governance and version-controlled, fully automated pipelines from day one.
- If your primary focus is rapid prototyping: Start with a small, manually curated dataset to validate your AI approach before investing in a full-scale pipeline.
- If your primary focus is scaling across a large organization: Invest heavily in centralized data storage, shared ontologies, and common pipeline components to prevent data silos.
Ultimately, treating your data with the same rigor as your wet lab experiments is the foundation of building successful and reliable biological AI.
Summary Table:
| Step | Key Action | Purpose |
|---|---|---|
| Data Governance | Establish naming conventions, ontologies, metadata standards | Ensure consistency and comparability across experiments |
| Data Pipeline | Ingest, parse, QC, normalize, extract features, assemble | Automate transformation of raw data into AI-ready feature matrix |
| Trade-offs | Balance over-processing vs. under-processing, manage overhead | Optimize for model performance and avoid data leakage |
Struggling to standardize your wet lab data for AI? KINTEK specializes in lab press machines, including automatic lab presses, isostatic presses, and heated lab presses, serving laboratories to enhance data reliability and experimental efficiency. Let us help you achieve consistent results—contact us today to discuss your needs and discover how our solutions can support your AI-driven research!
Visual Guide
Related Products
- Manual Heated Hydraulic Lab Press with Integrated Hot Plates Hydraulic Press Machine
- Laboratory Hydraulic Press 2T Lab Pellet Press for KBR FTIR
- Automatic Heated Hydraulic Press Machine with Hot Plates for Laboratory
- Laboratory Manual Heated Hydraulic Press Machine with Hot Plates
- Heated Hydraulic Press Machine with Heated Plates for Vacuum Box Laboratory Hot Press
People Also Ask
- Why is high-precision temperature control of laboratory hydraulic press heating plates critical for wood densification?
- What is the role of a hydraulic press with heating capabilities in constructing the interface for Li/LLZO/Li symmetric cells? Enable Seamless Solid-State Battery Assembly
- What are the applications of hydraulic heat presses in material testing and research? Enhance Precision and Reliability in Your Lab
- How is the temperature of the hot plate controlled in a hydraulic lab press? Achieve Thermal Precision (20°C-200°C)
- How is a heated hydraulic press used in material testing and research? Unlock Precision in Material Analysis



















